In this tutorial we look at alphabets and you will learn how to work with nucleotides and amino acids in BioC++. We guide you through the most important properties of BioC++'s alphabets and show you the different implemented types. After completion, you will be able to use the alphabets inside of STL containers and to compare alphabet values.
| Difficulty | Easy |
|---|---|
| Duration | 45 min |
| Prerequisite tutorials | Quick Setup (using CMake) |
| Recommended reading | None |
The links on this page mostly point straight into the API documentation which you should use as a reference. The code examples and assignments are designed to provide some practical experience with our interface as well as a code basis for your own program development.
An alphabet is the set of symbols of which a biological sequence – or in general a text – is composed. BioC++ implements specific and optimised alphabets not only for sequences of RNA, DNA and protein components, but also for quality, secondary structure and gap annotation as well as combinations of the aforementioned.
Nucleotides are the components of (Deoxy)Ribonucleic acid (DNA/RNA) and contain one of the nucleobases Adenine (A), Cytosine (C), Guanine (G), Thymine (T, only DNA) and Uracil (U, only RNA). In BioC++ the alphabets bio::alphabet::dna4 and bio::alphabet::rna4 contain exactly the four respective nucleotides. The trailed number in the alphabets' name represents the number of entities the alphabet holds – we denote this number as alphabet size. For instance, the alphabet bio::alphabet::dna5 represents five entities as it contains the additional symbol 'N' to refer to an unknown nucleotide.
Let's look at some example code which demonstrates how objects of the bio::alphabet::dna4 alphabet are assigned from characters.
We have shown three solutions for assigning variables of alphabet type.
_dna4 to the respective char symbol. char via the global function bio::alphabet::assign_char. The rank of a symbol is a number in range [0..alphabet_size) where each number is paired with an alphabet symbol by a bijective function. In BioC++ the rank is always determined by the lexicographical order of the underlying characters. For instance, in bio::alphabet::dna4 the bijection is
'A'_dna4 ⟼ 0
'C'_dna4 ⟼ 1
'G'_dna4 ⟼ 2
'T'_dna4 ⟼ 3.
BioC++ provides the function bio::alphabet::to_rank for converting a symbol to its rank value as demonstrated in the following code example. Note that the data type of the rank is usually the smallest possible unsigned type that is required for storing the values of the alphabet.
Our alphabets also have a character representation because it is more intuitive to work with them than using the rank. Each alphabet symbol is represented by its respective character whenever possible (A ⟼ 'A'). Analogously to the rank, BioC++ provides the function bio::alphabet::to_char for converting a symbol to its character representation.
Above you have seen that you can assign an alphabet symbol from a character with bio::alphabet::from_char. In contrast to the rank interface, this assignment is not a bijection because the whole spectrum of available chars is mapped to values inside the alphabet. For instance, assigning to bio::alphabet::dna4 from any character other than C, G or T results in the value 'A'_dna4 and assigning from any character except A, C, G or T to bio::alphabet::dna5 results in the value 'N'_dna5. You can avoid the implicit conversion by using bio::alphabet::assign_char_strict which throws bio::alphabet::invalid_char_assignment on invalid characters.
You can test the validity of a character by calling bio::alphabet::char_is_valid_for. It returns true if the character is valid and false otherwise.
You can retrieve the alphabet size by accessing the class member variable alphabet_size which is implemented in most bio::alphabet::alphabet instances.
In BioC++ you can use the STL containers to model e.g. sequences, sets or mappings with our alphabets. The following example shows some exemplary contexts for their use. For sequences we recommend the std::vector with one of BioC++'s alphabet types. Please note how easily a sequence can be created via the string literal.
All alphabets in BioC++ are regular and comparable. This means that you can use the <, <=, > and >= operators to compare the values based on the rank. For instance, this is useful for sorting a text over the alphabet. Regular implies that the equality and inequality of two values can be tested with == and !=.
To wrap up this section on the nucleotide alphabet, the following assignment lets you practise the use of a BioC++ alphabet and its related functions. It will also show you a handy advantage of using a vector over an alphabet instead of using std::string: The rank representation can be used straight as an array index (opposed to using a map with logarithmic access times, for example).
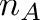
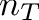
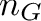
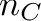
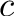
![\[ c = \frac{n_G + n_C}{n_A + n_T + n_G + n_C} \]](form_11.png)
Write a program that
alphabet size andThe bio::alphabet::dna5 type ensures that invalid characters in the input sequence are converted to 'N'. Note that these characters should not influence the GC content.
Pass the sequences CATTACAG (3/8 = 37.5%) and ANNAGAT (1/5 = 20%) to your program and check if your results are correct.
Until now, we have focused on alphabets for nucleotides to introduce the properties of BioC++'s alphabet on a specific example. BioC++ implements, however, many more alphabets. In this section, we want to give you an overview of the other existing alphabets.
Proteins consist of one or more long chains of amino acids. The so-called primary structure of a protein is expressed as sequences over an amino acid alphabet. The bio::alphabet::aa27 alphabet contains the standard one-letter code of the 20 canonical amino acids as well as the two proteinogenic amino acids, a termination symbol and some wildcard characters. For details read the Aminoacid page.
The alphabets for quality are sequence annotations since they describe additional properties of the respective sequence. The values are produced by sequencing machines and represent the probability that a nucleobase was recorded incorrectly. The characters are most commonly found in FASTQ files. See Quality for details.
You can build an Alphabet Tuple Composite with a nucleotide and quality alphabet that stores both information together. For the use cases just described we offer a pre-defined composite (bio::alphabet::qualified). See our API documentation for a detailed description of each.
The bio::alphabet::gap alphabet is the smallest alphabet in BioC++, consisting only of the gap character. It is most often used in an Alphabet Variant with a nucleotide or amino acid alphabet to represent gapped sequences, e.g. in alignments. To create a gapped alphabet simply use bio::alphabet::gapped<> with the alphabet type you want to refine.