Provides the various quality score types. More...
 Collaboration diagram for Quality:
Collaboration diagram for Quality:Classes | |
| class | bio::alphabet::phred42 |
| Quality type for traditional Sanger and modern Illumina Phred scores (typical range).. More... | |
| class | bio::alphabet::phred63 |
| Quality type for traditional Sanger and modern Illumina Phred scores (full range).. More... | |
| class | bio::alphabet::phred68legacy |
| Quality type for Solexa and deprecated Illumina formats.. More... | |
| class | bio::alphabet::qualified< sequence_alphabet_t, quality_alphabet_t > |
| Joins an arbitrary alphabet with a quality alphabet. More... | |
| class | bio::alphabet::quality_base< derived_type, size > |
| A CRTP-base that refines bio::alphabet::base and is used by the quality alphabets. More... | |
Concepts | |
| concept | bio::alphabet::quality |
| A concept that indicates whether an alphabet represents quality scores. | |
| concept | bio::alphabet::writable_quality |
| A concept that indicates whether a writable alphabet represents quality scores. | |
Typedefs | |
| template<typename alphabet_type > | |
| using | bio::alphabet::phred_t = decltype(bio::alphabet::to_phred(std::declval< alphabet_type >())) |
The phred_type of the alphabet; defined as the return type of bio::alphabet::to_phred. | |
Function objects (Quality) | |
| constexpr auto | bio::alphabet::to_phred |
| The public getter function for the phred representation of a quality score. | |
| constexpr auto | bio::alphabet::assign_phred_to |
| Assign a phred score to a quality alphabet object. | |
Provides the various quality score types.
Quality score sequences are usually output together with the DNA (or RNA) sequence by sequencing machines like the Illumina Genome Analyzer. The quality score of a nucleotide is also known as Phred score and is an integer score being inversely proportional to the propability 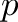
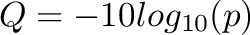
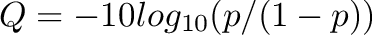
| Format | Quality Type | Phred Score Range | Rank Range | ASCII Range | Assert |
|---|---|---|---|---|---|
| Sanger, Illumina 1.8+ short | bio::alphabet::phred42 | [0 .. 41] | [0 .. 41] | ['!' .. 'J'] | Phred score in [0 .. 61] |
| Sanger, Illumina 1.8+ long | bio::alphabet::phred63 | [0 .. 62] | [0 .. 62] | ['!' .. '_'] | Phred score in [0 .. 62] |
| Solexa, Illumina [1.0; 1.8[ | bio::alphabet::phred68legacy | [-5 .. 62] | [0 .. 67] | [';' .. '~'] | Phred score in [-5 .. 62] |
The most distributed format is the Sanger or Illumina 1.8+ format. Despite typical Phred scores for Illumina machines range from 0 to maximal 41, it is possible that processed reads reach higher scores. If you don't intend to handle Phred scores larger than 41, we recommend to use bio::alphabet::phred42 due to its more space efficient implementation. For other formats, like Solexa and Illumina 1.0 to 1.7 the type bio::alphabet::phred68legacy is provided. To cover also the Solexa format, the Phred score is stored as a signed integer starting at -5. An overview of all the score formats and their encodings can be found here: https://en.wikipedia.org/wiki/FASTQ_format#Encoding.
The quality submodule defines the bio::alphabet::writable_quality which encompasses all the alphabets, defined in the submodule, and refines the bio::alphabet::writable_alphabet by providing Phred score assignment and conversion operations. Additionally, this submodule defines the bio::alphabet::quality, which only requires readablity and not assignability.
Quality alphabets can be converted to their char and rank representation via bio::alphabet::to_char and bio::alphabet::to_rank respectively (like all other alphabets). Additionally they can be converted to their Phred representation via bio::alphabet::to_phred.
Likewise, assignment happens via bio::alphabet::assign_char_to, bio::alphabet::assign_rank_to and bio::alphabet::assign_phred_to. Phred values outside the representable range, but inside the legal range, are converted to the closest Phred score, e.g. assigning 60 to a bio::alphabet::phred42 will result in a Phred score of 41. Assigning Phred values outside the legal range results in undefined behaviour.
All quality alphabets are explicitly convertible to each other via their Phred representation. Values not present in one alphabet are mapped to the closest value in the target alphabet (e.g. a bio::alphabet::phred63 letter with value 60 will convert to a bio::alphabet::phred42 letter of score 41).
|
inlineconstexpr |
Assign a phred score to a quality alphabet object.
| alph_type | The type of the target object. Must model the bio::alphabet::quality. |
| phr | The phred score being assigned; must be of the bio::alphabet::phred_t of the target object. |
alph if alph was given as lvalue, otherwise a copy.This is a function object. Invoke it with the parameter(s) specified above.
It is defined for all quality alphabets in BioC++.
This is a customisation point (see Customisation). If you don't want to create your own alphabet, everything below is irrelevant to you!
This object acts as a wrapper and looks for an implementation with the following signature:
Functions are found via ADL and considered only if they are marked noexcept (constexpr is not required, but recommended) and if the returned type is exactly alph_type &.
To specify the behaviour for your own alphabet type, simply provide the above function as a friend or free function.
Note that temporaries of alph_type are handled by this function object and do not require an additional overload.
|
inlineconstexpr |
The public getter function for the phred representation of a quality score.
| your_type | The type of alphabet. Must model the bio::alphabet::quality. |
| chr | The quality value to convert into the phred score. |
This is a function object. Invoke it with the parameter(s) specified above.
It is defined for all quality alphabets in BioC++.
This is a customisation point (see Customisation). If you don't want to create your own alphabet, everything below is irrelevant to you!
This object acts as a wrapper and looks for an implementation with the following signature:
Functions are found via ADL and considered only if they are marked noexcept (constexpr is not required, but recommended) and if the returned type models std::integral.
To specify the behaviour for your own alphabet type, simply provide the above function as a friend or free function.